The breast cancer world is somewhat fortunate to have biomarkers that are great at selecting patients most likely to respond to targeted therapies. The stratification of breast cancer into estrogen receptor–positive (ER+) and ER-negative disease has stood the test of time. The identification of HER2 and the development of HER2-directed monoclonal antibodies has resulted in the recognition of another important therapeutic subtype of breast cancer. ER and HER2 are good selection markers for therapeutics; however, not all patients with positive status will respond to therapy. The quest for biomarkers predictive of response has led to the realization of the importance of proliferation pathways in ER+ cancers. Most gene signatures for ER+ cancers use proliferation-related genes, in addition to others, for the stratification of patients to determine the likelihood of recurrence and more important the need for chemotherapy. Unfortunately, the same has not been true for tumors overexpressing HER2 and for tumors lacking expression of estrogen (progesterone) and HER2 receptors (triple-negative breast cancers).
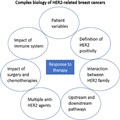
The challenge of developing gene signatures for HER2-positive (HER2+) cancers seemed daunting as a number of mechanisms in clinical and experimental models, have been shown to be important (Figure 1) [see Swain et al.[1] for a recent review]. The first and foremost of these was the definition of HER2 positivity. There is a high but not perfect correlation concordance between immunohistochemical assays and between immunohistochemical and fluorescent in situ hybridization and mRNA levels.[2–5] The HER2 pathway is complex with the formation of homo- and heterodimers with other family members.[1] Dual targeting of HER2 by trastuzumab and pertuzumab results in an improved response. The HER2 and HER3 heterodimer has been implicated in therapeutic resistance. Targeting the downstream effectors such as Phosphatidylinositol 3-Kinase (PI3k), AKT (also known as protein kinase B) and mammalian target of rapamycin (mTOR) pathways has been associated with therapeutic benefit. An antibody-drug conjugate against HER3 has been recently developed and shown to have therapeutic efficacy. However, the benefit did not seem to depend on the levels of HER3 protein. Additional studies are clearly necessary to elucidate the roles of HER2 and its family members in determination of response.
Figure 1.
Complex biology of HER2-related breast cancers. Response to therapy is multifactorial and impacted by patient variable, HER2 expression, and its interaction with other molecular pathways as well as by the therapeutic regimens prescribed for the patient.
Accumulating evidence has documented a prognostic role of tumor-infiltrating lymphocytes in HER2+ cancers. Tumor-infiltrating lymphocytes are associated with better survival and higher likelihood of obtaining a complete pathological response (pCR).[6–8] A number of gene expression signatures that quantitate immune response have also been described. The expression of checkpoint inhibitors such as programmed death ligand–1 has also been assessed in HER2+ tumors and has resulted in several clinical trials studying the combination of immune and HER2-directed therapies.[1] The importance of immune mechanisms is also highlighted by the role of fragment crystallizable gamma receptor (FcγR)–dependent activities, which include induction of antibody-dependent cellular cytotoxicity and antibody-dependent cellular phagocytosis, in cancer cell elimination.[9] Margetuximab, a US Food and Drug Administration–approved anti-HER2 Fc-engineered chimeric mAb, targets the same epitope as trastuzumab but has increased binding to activating FcγRIIIa and decreased binding to inhibitory FcγRIIb (CD32B).[10–12]
In addition to the complexities discussed above, one should bear in mind that most anti-HER2 therapeutic trials are in a background of standard of care therapies including surgery and multiagent chemotherapy. The benefit observed in a (but clinically significant) fraction of patients could be because of 1 or more of these therapies (ie, you can cure a patient only once!).
This tangled web of biological and therapeutic factors has presented a formidable barrier to the development of a gene signature to predict risk of recurrence or response to therapy in HER2+ breast cancer. Prat and colleagues[13] seem to have cracked this barrier. They have developed a gene signature using a slightly unusual approach. Rather than using the classical unsupervised approach to identifying key genes associated with adverse outcomes, they have used prior knowledge to develop a "handcrafted" supervised approach. They have recently described "HER2D" signature to predict the outcomes of patients treated with chemotherapy and trastuzumab.[13,14] The signature incorporates tumor size, nodal status, and 27 genes representing 4 gene expression signatures related to immune infiltration, proliferation, tumor cell differentiation, and HER2 amplicon. They developed 2 models: 1) a prognostic model and 2) a predictor of likelihood of pCR after anti-HER2–based chemotherapy. The prognostic risk-score model was developed using a series of 434 patients and validated in an independent cohort of 268 patients in addition to 5 publicly available datasets. The model performed very well in the validation set for the prediction of disease-free survival (P = .002) with the 5-year survival in the low-risk group of 97.4%. The pCR model was developed using 116 patients and validated in 2 independent cohort (n = 91 and 67 cases). The continuous pCR model, similarly, was statistically significantly associated with pCR (P < .0001). Interestingly, the 2 models—risk score and pCR score—showed only a weak relationship (correlation coefficient = -0.19). In another study, Guarneri et al.[14] showed that HER2DX could predict response following neoadjuvant letrozole in combination with dual HER2 blockade with trastuzumab and pertuzumab in early stage HER2+ and hormone receptor–positive breast cancer. The limitations of these studies were that they were based on patients with early stage disease treated with trastuzumab-based therapies.
In this issue of the Journal, Brasó-Maristany et al.[15] seek to address some of these limitations and document utility of the risk-score patients with advanced disease treated with trastuzumab emtansine (T-DM1), an antibody drug conjugate. In contrast to the prior work, they find a relatively weak but still statistically significant association (P = .04) of the risk score with overall survival in multivariable analyses. The observed result could be due to a combination of factors including small sample size of the study (n = 91). The patient population is also heterogeneous, with 46% of the patients having de novo metastatic disease. The remainder of the patients had 1 or more rounds of prior therapies. The HER2Dx assay was also performed on archival samples and includes tissue from primary as well as metastatic sites. Overall, the data have the promise of having utility in a bigger, better controlled patient population.
Brasó-Maristany et al.[15] also study the importance of HER2 mRNA, 1 of the 27 genes in the HER2Dx assay. Low levels of mRNA are found in approximately 10% of HER2+ cases. This is prima facie contrary to the expectation that all clinically HER2+ patients will have high mRNA. However, this is in fact consistent with prior data. Perez et al.[16] studied the correlation of HER2 mRNA performed using 21-gene recurrence score assay and had noted that approximately 14% of the 901 cases analyzed from the N9831 clinical trial had low mRNA levels. Perez et al.[16] also observed only a trend for mRNA levels to correlate with response to trastuzumab (P = .057). In contrast, in the T-DM1–based EMILIA trial, there was a survival benefit of T-DM1 was greater than compared with a capecitabine plus lapatinib regimen in patients with high levels of HER2 mRNA.[17] A similar result was observed in the TH3RESA clinical trial.[18] Last, in the MARIANNE clinical trial, HER2 mRNA levels greater than median were associated with numerically longer progression-free survival in all arms of the trial.[19] Collectively, these data substantiate the observations made by Brasó-Maristany et al.,[15] who find a strong correlation of the mRNA levels with overall response to T-DM1 (0% in low, 29% in intermediate, and 56% in high). It is important to note that the current study was performed on archival primary as well as metastatic samples. Although variability in HER2 protein expression in archival tissues has been studied, the variability of HER2 mRNA in old tissues is not well documented . The authors state that based on prior experience,[20] they did not expect mRNA variability to be a major issue.
The main question that arises now is, where does the HER2DX assay fit in to today's practice? The landscape of HER2+ cancers is rapidly evolving with the advent of novel anti-HER2 agents. The results of the DESTINY-04 clinical trial show that trastuzumab deruxtecan is a potent agent even in patients with low levels of HER2 expression (1+ or 2+; fluorescent in situ hybridization–negative).[21] A number of other agents such as tyrosine kinase inhibitors (eg, US Food and Drug Administration–approved neratinib and tucatinib) and other novel antibody drug conjugates are also available.[1] It would be interesting to see if HER2Dx could help stratify this increasingly complex landscape and provide patients and their clinicians a tool to personalize and select the most appropriate anti-HER2 agent for therapeutics. The assay could also help sequence these agents to optimize therapeutic response.
In conclusion, the work by Brasó-Maristany et al.[15] not only builds on the prior data regarding HER2 mRNA levels and likelihood of response but also supports the further development of the HER2DX genomic assay. This is likely to become the go-to assay for assessment of risk of progression in patients with early- and advanced-stage HER2+ disease and predicting the likelihood of response to neoadjuvant chemotherapy.
Funding
No funding was used for this editorial.
Notes
Role of the funder
Not applicable.
Acknowledgements
Drs Badve and Gökmen-Polar are supported by Funds from Department of Pathology, Emory University, and from Susan G Komen leadership grant (SAC220219) to SSB.
Disclaimers
The thoughts and opinions expressed in this editorial are the author's own and do not reflect those of Oxford University Press or Emory University.
Data availability
No new data were generated or analyzed for this editorial.
J Natl Cancer Inst. 2023;115(3):242-245. © 2023 Oxford University Press